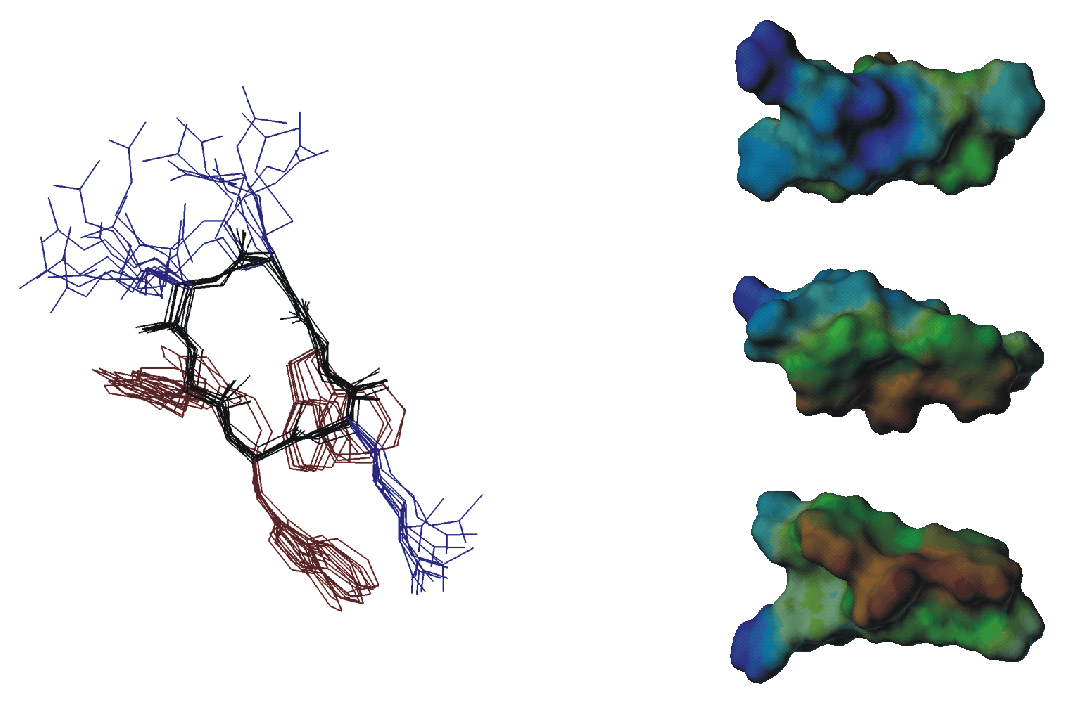
The widespread phenomenon of multidrug resistance in bacteria has necessitated the search for new antimicrobial substances. About 500 naturally occuring antimicrobial peptides are known that could provide a starting point for the development of new drugs. They exihibit a large variety of sequences, secondary structures and contain L- as well as D- and unnatural amino acids. Several hypotheses regarding their mechanism of action have been presented. The goal of this project is to obtain three-dimensional structures of small anitmicrobial peptides in a membrane mimicking environment using solution state NMR since only with structural information at hand a detailed picture of the mechanism can be developed.