deposited 20.12.2018
released
29.01.2020
Peter Schmieder: Structures
| 6QAY | 5KGQ | 5NCA | 2MWG | 2MBK | |||||
| 2RQK | 2RQM | 2KD3 | 2OTQ | 2OX2 | 2JRZ | 2I9S | 2JNU | 2OWI | 2I59 |
| 2JM5 | 2EXG | 1XZ9 | 1R4T | 1SKI | 1SKK | 1SKL | 1S3A | 1QVK | 1QVL |
| 1OQA | 1OYI | 1R84 | 1R2N | 1PQS | 1EGX | 1QGP | 1SGG | 1RAX | 2PDZ |
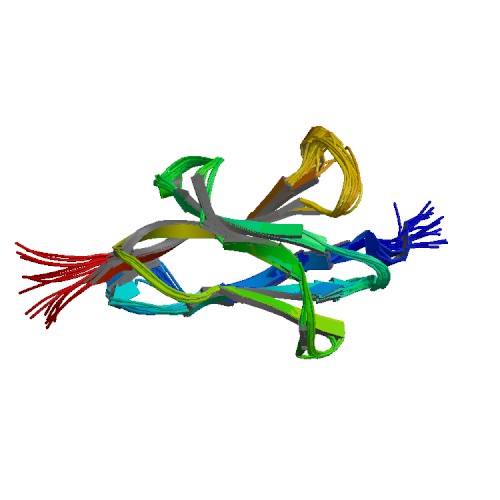
| 6QAY | Structural investigation of the TasA anchoring protein TapA from Bacillus subtilis |
 deposited 20.12.2018 |
Authors: Higman, V.; Schmieder, P.; Diehl, A.; Oschkinat, H. |
| Citation: | |
| 35 |
| 5KGQ | NMR structure and dynamics of Q4DY78, a conserved kinetoplasid-specific protein from Trypanosoma cruzi |
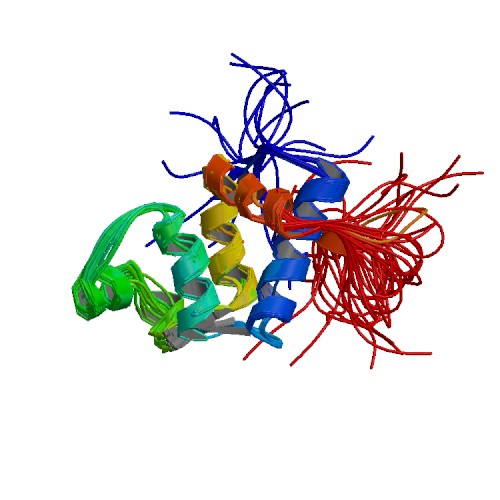 deposited 20.12.2018 |
Authors: D'Andrea, E.D., Retel, J.S., Diehl, A., Schmieder, P., Oschkinat, H., Pires, J.R. |
| Citation: | |
| 34 |
| 5NCA | Solution structure of ComGC from Streptococcus pneumoniae |
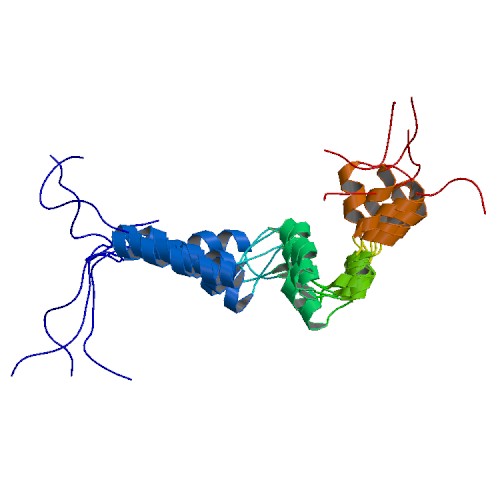 deposited 03.03.2017 |
Authors: Akbey, U. Aschtgen, M.S. Boesen, T. de Lichtenberg, C. Erlendsson, S. Henriques-Normark, B. Muschiol, S. Oliveira, V. Schmieder, P. Teilum, K. |
| Citation:S. Muschiol*; S. Erlendsson; M.S. Aschtgen; V. Oliveira; P. Schmieder; C. de Lichtenberg; K. Teilum; T. Boesen; U. Akbey; B. Henriques-Normark*; "Structure of the competence pilus major pilin ComGC in Streptococcus pneumoniae"; J. Biol. Chem. 292, 14134 - 14149 (2017) | |
| 33 |
| 2MWG | Full-Length Solution Structure Of YtvA, a LOV-Photoreceptor Protein and Regulator of Bacterial Stress Response |
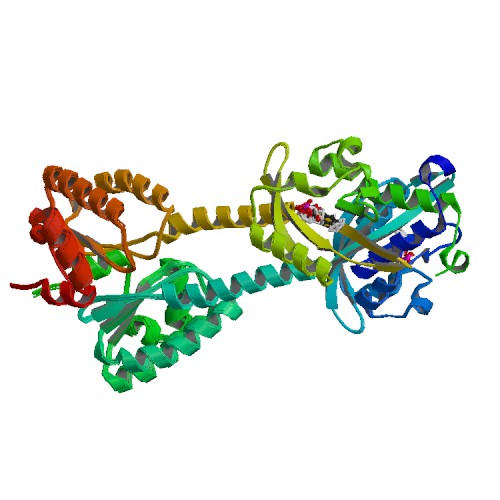 deposited 07.11.2014 |
Authors: Jurk, M., Bardiaux, B., Schmieder, P. |
| Citation: | |
| 32 |
| top |
| 2MBK | The Clip-segment of the von Willebrand domain 1 of the BMP modulator protein Crossveinless 2 is preformed |
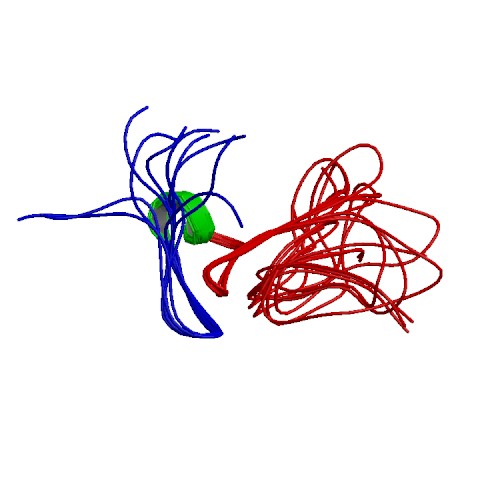 deposited 02.08.2013 |
Authors: Mueller, T.D., Fiebig, J.E., Weidauer, S.E., Qiu, L., Bauer, M., Schmieder, P., Beerbaum, M., Zhang, J., Oschkinat, H., Sebald, W. |
| Citation: J.E. Fiebig; S.E. Weidauer; L. Qiu; M. Bauer; P. Schmieder; M. Beerbaum; J. Zhang; H. Oschkinat; W. Sebald; T.D. Mueller*; "The Clip-segment of the von Willebrand domain 1 of the BMP modulator protein Crossveinless 2 is preformed"; Molecules 18, 11658-11682 (2013) | |
| 31 |
| 2RQK | NMR Solution Structure of Mesoderm Development (MESD) - closed conformation |
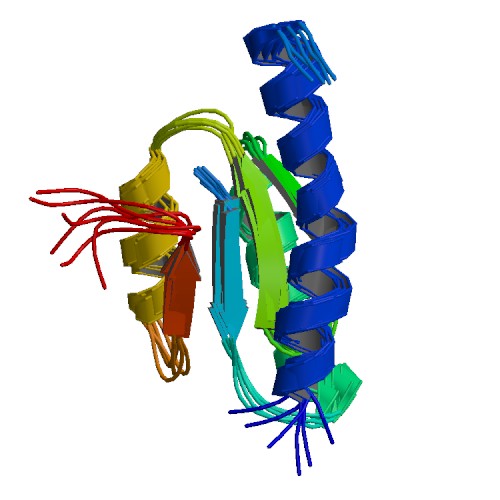 deposited 06.08.2009 |
Authors: Köhler, C., Lighthouse, J.K., Werther, T., Andersen, O.M., Diehl, A., Schmieder, P., Holdener, B.C., Oschkinat, H. |
| Citation: C. Köhler; J.K. Lighthouse; T. Werther; O.M. Andersen; A. Diehl; P. Schmieder; J. Du; B.C. Holdener; H. Oschkinat*; "The Structure of MESD45-184 Brings Light into the Mechanism of LDLR Family Folding"; Structure 19, 337-348 (2011) | |
| 30 |
| 2RQM | NMR Solution Structure of Mesoderm Development (MESD) - open conformation |
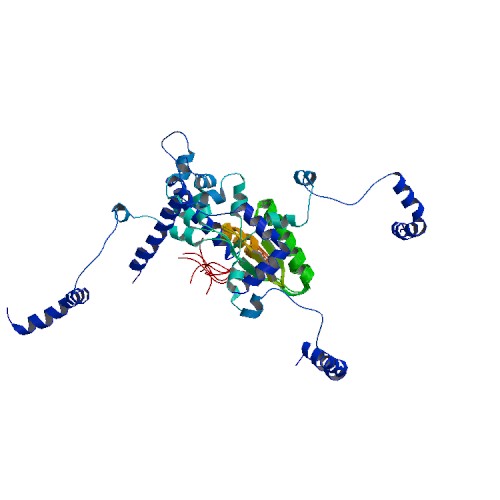 deposited 06.08.2009 |
Authors: Köhler, C., Lighthouse, J.K., Werther, T., Andersen, O.M., Diehl, A., Schmieder, P., Holdener, B.C., Oschkinat, H. |
| Citation: C. Köhler; J.K. Lighthouse; T. Werther; O.M. Andersen; A. Diehl; P. Schmieder; J. Du; B.C. Holdener; H. Oschkinat*; "The Structure of MESD45-184 Brings Light into the Mechanism of LDLR Family Folding"; Structure 19, 337-348 (2011) | |
| 29 |
| 2KD3 | NMR structure of the Wnt modulator protein Sclerostin |
 deposited 06.08.2009 |
Authors: Beerbaum, M. Mueller, T.D. Oschkinat, H. Schmieder, P. Schmitz, W. Weidauer, S.E. |
| Citation: S.E. Weidauer; P. Schmieder; M. Beerbaum; W. Schmitz; H. Oschkinat; T.D. Müller*; "NMR structure of the Wnt modulator protein Sclerostin"; Biochem. Biophys. Res. Commun. 380, 160-165 (2009) | |
| 28 |
| top |
| 2OTQ | Structure of the antimicrobial peptide cyclo(RRWFWR) bound to DPC micelles |
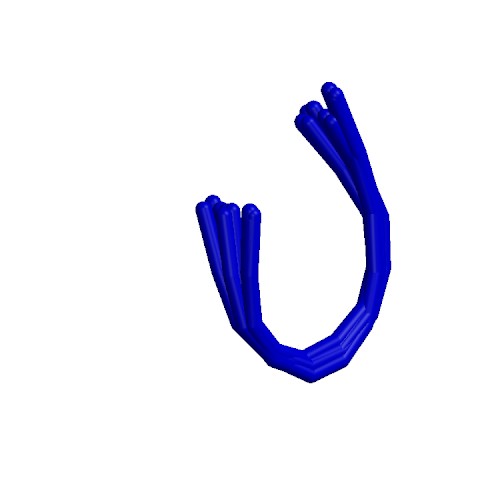 deposited 09.02.2007 |
Authors: Appelt, C., Wesselowski, A., Soderhall, J.A., Dathe, M., Schmieder, P. |
| Citation: C. Appelt; A. Wessolowski; M. Dathe; P. Schmieder*; "Structures of cyclic, antimicrobial peptides in a membrane-mimicking environment define requirements for activity"; J. Pept. Sci. 14, 524-527 (2008) | |
| 27 |
| 2OX2 | Structure of the antimicrobial peptide cyclo(RRWFWR) bound to DPC micelles |
 deposited 09.02.2007 |
Authors: Appelt, C., Wesselowski, A., Soderhall, J.A., Dathe, M., Schmieder, P. |
| Citation: C. Appelt; A. Wessolowski; M. Dathe; P. Schmieder*; "Structures of cyclic, antimicrobial peptides in a membrane-mimicking environment define requirements for activity"; J. Pept. Sci. 14, 524-527 (2008) | |
| 26 |
| 2JRZ | Solution structure of the Bright/ARID domain from the human JARID1C protein |
 deposited 29.06.2007 |
Authors: Köhler, C., Bishop, S., Dowler, E.F., Diehl, A., Schmieder, P., Leidert, M., Sundstrom, M., Arrowsmith, C.H., Wiegelt, J., Edwards, A., Oschkinat, H., Ball, L.J. |
| Citation: C. Koehler; S. Bishop; E. F. Dowler; P. Schmieder; A. Diehl; H. Oschkinat; L.J. Ball; "Backbone and sidechain 1H, 13C and 15N resonance assignments of the Bright/ARID domain from the human JARID1C (SMCX) protein" Biomol. NMR Assign. 2, 9-11 (2008) | |
| 25 |
| 2I9S | The solution structure of the core of mesoderm development (MESD) |
 deposited 06.09.2006 |
Authors: Köhler, C., Andersen, O., Diehl, A., Schmieder, P., Krause, G., Oschkinat, H |
| Citation: C. Köhler; O.M. Andersen; A. Diehl; G. Krause; P. Schmieder; H. Oschkinat*; "The solution structure of the core of mesoderm development (MESD), a chaperone for members of the LDLR-family"; J. Struct. Funct. Genomics 7, 131-138 (2006) | |
| 24 |
| top |
| 2JNU | Solution structure of the RGS domain of human RGS14 |
 deposited 02.02.2007 |
Authors: Dowler, E.F., Diehl, A., Bray, J., Elkins, J., Soundararajan, M., Doyle, D.A., Gileadi, C., Phillips, C., Schoch, G.A., Yang, X., Brockmann, C., Leidert, M., Rehbein, K., Schmieder, P., Kühne, R., Higman, V.A., Sundstrom, M., Arrowsmith, C., Weigelt, J., Edwards, A., Oschkinat, H., Ball, L.J |
| Citation: M. Soundararajan; F.S. Willard; A.J. Kimple; A.P. Turnbull; L.J. Ball; G.A. Schoch; C. Gileadi ; O.Y. Fedorov; E.F. Dowler; V.A. Higman; S.Q. Hutsell; M. Sundström; D.A. Doyle; D.P. Siderovski; "Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits"; Proc. Natl. Acad. Sci. USA 105, 6457-6462 (2008) | |
| 23 |
| 2OWI | Solution structure of the RGS domain from human RGS18 |
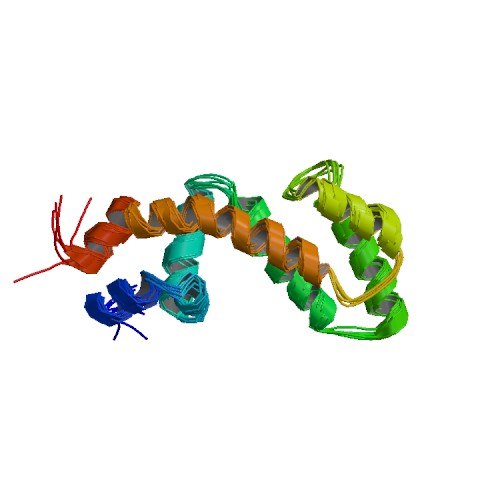 deposited 16.02.2007 |
Authors: Higman, V.A., Leidert, M., Bray, J., Elkins, J., Soundararajan, M., Doyle, D.A., Gileadi, C., Phillips, C., Schoch, G., Yang, X., Brockmann, C., Schmieder, P., Diehl, A., Sundstrom, M., Arrowsmith, C., Weigelt, J., Edwards, A., Oschkinat, H., Ball, L.J. |
| Citation: M. Soundararajan; F.S. Willard; A.J. Kimple; A.P. Turnbull; L.J. Ball; G.A. Schoch; C. Gileadi ; O.Y. Fedorov; E.F. Dowler; V.A. Higman; S.Q. Hutsell; M. Sundström; D.A. Doyle; D.P. Siderovski; "Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits"; Proc. Natl. Acad. Sci. USA 105, 6457-6462 (2008) | |
| 22 |
| 2I59 | Solution structure of RGS10 |
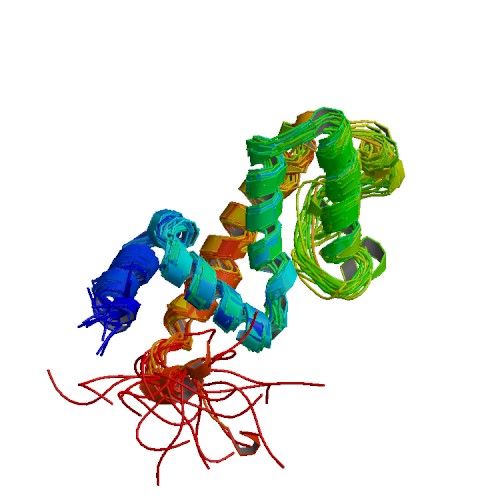 deposited 16.02.2007 |
Authors: Fedorov, O., Higman, V.A., Diehl, A., Leidert, M., Lemak, A., Schmieder, P., Oschkinat, H., Elkins, J., Soundarajan, M., Doyle, D.A., Arrowsmith, C., Sundstrom, M., Weigelt, J., Edwards, A., Ball, L.J., |
| Citation: M. Soundararajan; F.S. Willard; A.J. Kimple; A.P. Turnbull; L.J. Ball; G.A. Schoch; C. Gileadi ; O.Y. Fedorov; E.F. Dowler; V.A. Higman; S.Q. Hutsell; M. Sundström; D.A. Doyle; D.P. Siderovski; "Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits"; Proc. Natl. Acad. Sci. USA 105, 6457-6462 (2008) | |
| 21 |
| 2JM5 | Solution Structure of the RGS domain from human RGS18 |
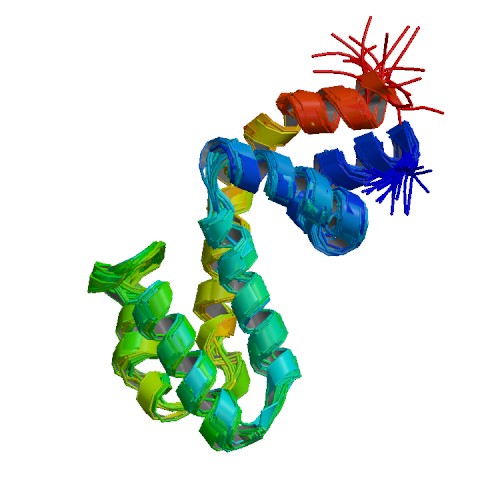 deposited 16.02.2007 |
Authors: Higman, V.A., Leidert, M., Bray, J., Elkins, J., Soundararajan, M., Doyle, D.A., Gileadi, C., Phillips, C., Schoch, G., Yang, X., Brockmann, C., Schmieder, P., Diehl, A., Sundstrom, M., Arrowsmith, C., Weigelt, J., Edwards, A., Oschkinat, H., Ball, L.J. |
| Citation: M. Soundararajan; F.S. Willard; A.J. Kimple; A.P. Turnbull; L.J. Ball; G.A. Schoch; C. Gileadi ; O.Y. Fedorov; E.F. Dowler; V.A. Higman; S.Q. Hutsell; M. Sundström; D.A. Doyle; D.P. Siderovski; "Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits"; Proc. Natl. Acad. Sci. USA 105, 6457-6462 (2008) | |
| 20 |
| top |
| 2EXG | Making Protein-Protein Interactions Drugable: Discovery of Low-Molecular-Weight Ligands for the AF6 PDZ Domain |
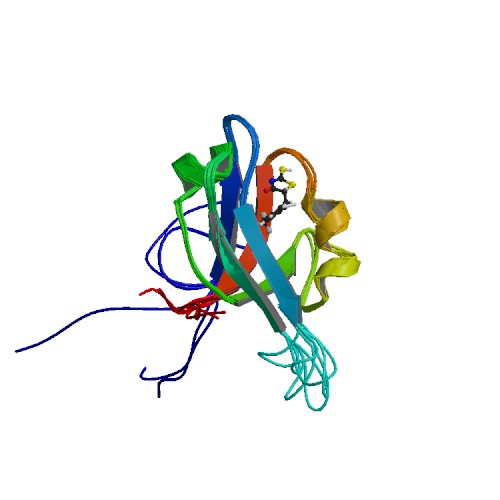 deposited 08.11.2005 |
Authors: Boisguerin, P. Diehl, A. Hagen, V. Joshi, M. Krause, G. Moelling, K. Oschkinat, H. Schade, M. Schmieder, P. Vargas, C. |
| Citation: M. Joshi; C. Vargas; P. Boisguerin; A. Diehl; G. Krause; P. Schmieder; K. Mölling; V. Hagen; M.Schade*; H. Oschkinat; "Discovery of low-molecular-weight ligands for the AF6 PDZ domain"; Angew. Chem. Int. Ed. Engl. 45, 3790-3795 (2006); Angew. Chem. 118, 3874–3879 (2006) | |
| 19 |
| 1XZ9 | Structure of AF-6 PDZ domain |
 deposited 12.11.2004 |
Authors: Joshi, M., Boisguerin, P., Leitner, D., Volkmer-Engert, R., Moelling, K., Schade, M., Schmieder, P., Krause, G., Oschkinat, H. |
| Citation: M. Joshi; C. Vargas; P. Boisguerin; A. Diehl; G. Krause; P. Schmieder; K. Mölling; V. Hagen; M.Schade*; H. Oschkinat; "Discovery of low-molecular-weight ligands for the AF6 PDZ domain"; Angew. Chem. Int. Ed. Engl. 45, 3790-3795 (2006); Angew. Chem. 118, 3874–3879 (2006) | |
| 18 |
| 1R4T | Solution structure of exoenzyme S |
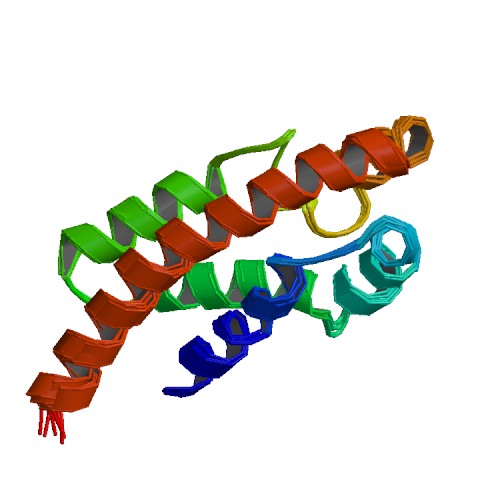 deposited 18.10.2003 |
Authors: Langdon, G.M., Leitner, D., Labudde, D., Kühne, R., Schmieder, P., Aktories, K., Oschkinat, H.O., Schmidt, G. |
| Citation: | |
| 17 |
| 1SKI | Structure of the antimicrobial hexapeptide cyc-(RRYYRF) bound to DPC micelles |
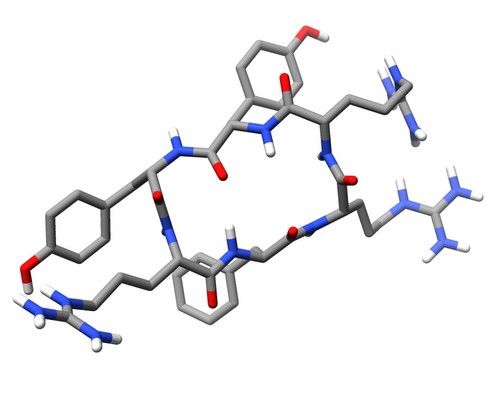 deposited 05.03.2004 |
Authors: Appelt, C., Soderhall, J.A., Bienert, M., Dathe, M., Schmieder, P. |
| Citation: C. Appelt; A. Wessolowski; J. A. Söderhäll; M. Dathe; P. Schmieder*; "Structure of the antimicrobial, cationic hexapeptide cyclo(RRWWRF) and its analogs in solution and bound to detergent micelles"; ChemBioChem 6, 1654-1662 (2005) | |
| 16 |
| top |
| 1SKK | Structure of the antimicrobial hexapeptide cyc-(KKWWKF) bound to DPC micelles |
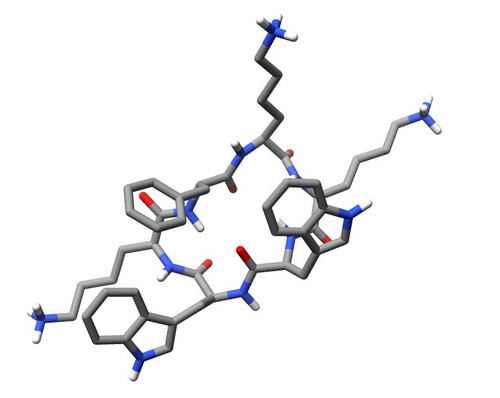 deposited 05.03.2004 |
Authors: Appelt, C., Soderhall, J.A., Bienert, M., Dathe, M., Schmieder, P. |
| Citation: C. Appelt; A. Wessolowski; J. A. Söderhäll; M. Dathe; P. Schmieder*; "Structure of the antimicrobial, cationic hexapeptide cyclo(RRWWRF) and its analogs in solution and bound to detergent micelles"; ChemBioChem 6, 1654-1662 (2005) | |
| 15 |
| 1SKL | Structure of the antimicrobial hexapeptide cyc-(RRNalNalRF) bound to DPC micelles |
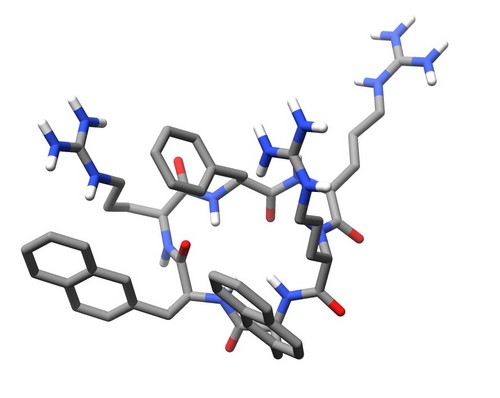 deposited 05.03.2004 |
Authors: Appelt, C., Soderhall, J.A., Bienert, M., Dathe, M., Schmieder, P. |
| Citation: C. Appelt; A. Wessolowski; J. A. Söderhäll; M. Dathe; P. Schmieder*; "Structure of the antimicrobial, cationic hexapeptide cyclo(RRWWRF) and its analogs in solution and bound to detergent micelles"; ChemBioChem 6, 1654-1662 (2005) | |
| 14 |
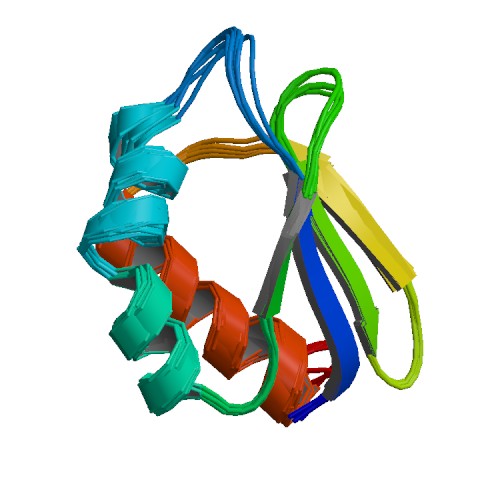
| 1S3A | NMR Solution Structure of Subunit B8 from Human NADH-Ubiquinone Oxidoreductase Complex I (CI-B8) |
 deposited 13.01.2004 |
Authors: Brockmann, C. Diehl, A. Korn, B. Kühne, R. Oschkinat, H. Rehbein, K. Schmieder, P. Strauss, H. |
| Citation: C. Brockmann; A. Diehl; K. Rehbein; H. Strauss; P. Schmieder; B. Korn; R. Kühne; H. Oschkinat*; "The oxidized subunit b8 from human complex I adopts a thioredoxin fold"; Structure 12, 1645-1654 (2004) | |
| 13 |
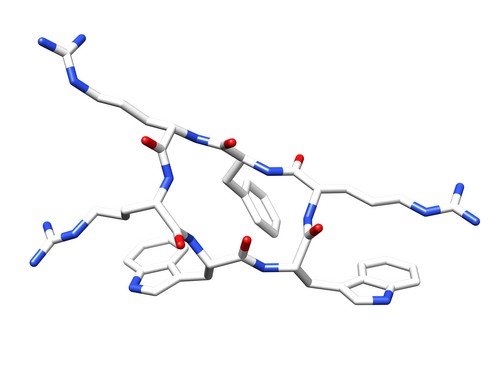
| 1QVK | Structure of the antimicrobial hexapeptide cyc-(RRWWRF) bound to DPC micelles |
 deposited 28.08.2003 |
Authors: Appelt, C., Soderhall, J.A., Bienert, M., Dathe, M., Schmieder, P. |
| Citation: C. Appelt; A. Wessolowski; J. A. Söderhäll; M. Dathe; P. Schmieder*; "Structure of the antimicrobial, cationic hexapeptide cyclo(RRWWRF) and its analogs in solution and bound to detergent micelles"; ChemBioChem 6, 1654-1662 (2005) | |
| 12 |
| top |
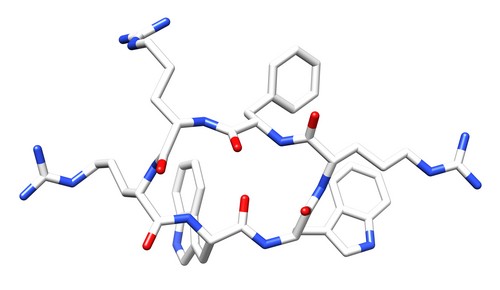
| 1QVL | Structure of the antimicrobial hexapeptide cyc-(RRWWRF) bound to SDS micelles |
 deposited 28.08.2003 |
Authors: Appelt, C., Soderhall, J.A., Bienert, M., Dathe, M., Schmieder, P. |
| Citation: C. Appelt; A. Wessolowski; J. A. Söderhäll; M. Dathe; P. Schmieder*; "Structure of the antimicrobial, cationic hexapeptide cyclo(RRWWRF) and its analogs in solution and bound to detergent micelles"; ChemBioChem 6, 1654-1662 (2005) | |
| 11 |
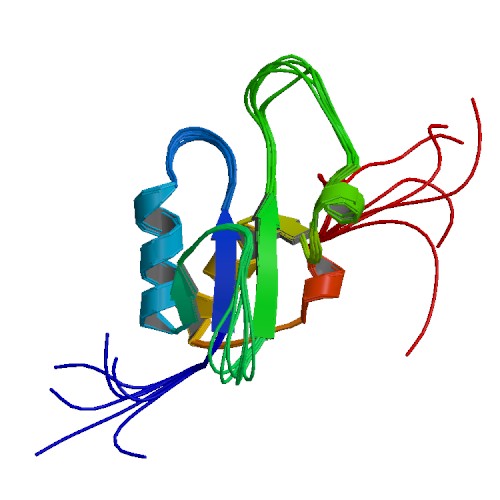
| 1OQA | Solution structure of the BRCT-c domain from human BRCA1 |
 deposited 07.03.2003 |
Authors: Gaiser, O.J., Ball, L.J., Schmieder, P., Leitner, D., Strauss, H., Wahl, M., Kühne, R., Oschkinat, H., Heinemann, U. |
| Citation: O.J. Gaiser; L.J. Ball*; P. Schmieder; D. Leitner; H. Strauss; M. Wahl; R. Kühne; H. Oschkinat; U. Heinemann; "Solution Structure, Backbone Dynamics, and Association Behavior of the C-Terminal BRCT Domain from the Breast Cancer-Associated Protein BRCA1"; Biochemistry 43, 15983-15995 (2004) | |
| 10 |
| 1OYI | Solution structure of the Z-DNA binding domain of the vaccinia virus gene E3L |
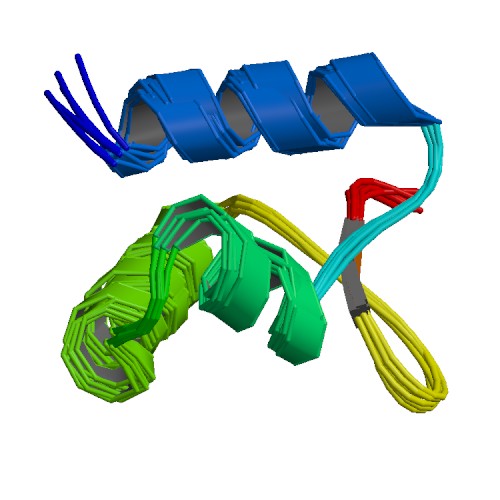 deposited 04.04.2003 |
Authors: Kahmann, J.D., Wecking, D.A., Putter, V., Lowenhaupt, K., Kim, Y.-G., Schmieder, P., Oschkinat, H., Rich, A., Schade, M. |
| Citation: J.D. Kahmann; D.A. Wecking; V. Putter; K. Lowenhaupt; Y.G. Kim; P. Schmieder; H. Oschkinat; A. Rich; M. Schade*; "The solution structure of the N-terminal domain of E3L shows a tyrosine conformation that may explain its reduced affinity to Z-DNA in vitro"; Proc. Natl. Acad. Sci. USA 101, 2712-2717 (2004) | |
| 09 |
| 1R84 | NMR structure of the 13-cis-15-syn retinal in dark_adapted bacteriorhodopsin |
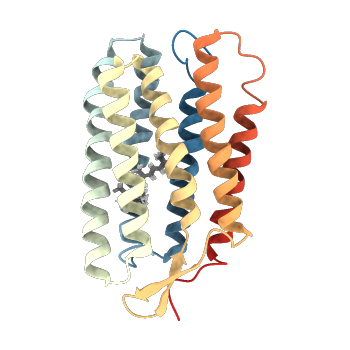 deposited 23.10.2003 |
Authors: Patzelt, H., Simon, B., Ter Laak, A., Kessler, B., Kühne, R., Schmieder, P., Oesterhaelt, D., Oschkinat, H. |
| Citation: H. Patzelt; B. Simon; A. terLaak; B. Kessler; R. Kühne; P. Schmieder; D. Oesterhelt*; H. Oschkinat*; “The structure of the active center in dark-adapted Bacteriorhodopsin by solution-state NMR spectroscopy”; Proc. Natl. Acad. Sci. USA 99, 9765-9770 (2002) | |
| 08 |
| top |
| 1R2N | NMR structure of the all-trans retinal in dark-adapted Bacteriorhodopsin |
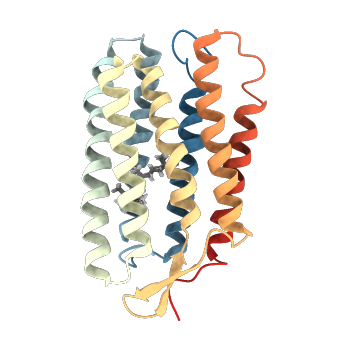 deposited 23.10.2003 |
Authors: Patzelt, H., Simon, B., Ter Laak, A., Kessler, B., Kühne, R., Schmieder, P., Oesterhaelt, D., Oschkinat, H. |
| Citation: H. Patzelt; B. Simon; A. terLaak; B. Kessler; R. Kühne; P. Schmieder; D. Oesterhelt*; H. Oschkinat*; “The structure of the active center in dark-adapted Bacteriorhodopsin by solution-state NMR spectroscopy”; Proc. Natl. Acad. Sci. USA 99, 9765-9770 (2002) | |
| 07 |
| 1PQS | Solution structure of the C-terminal OPCA domain of yCdc24p |
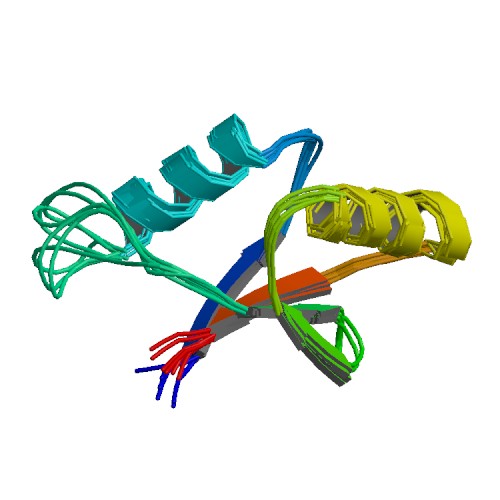 deposited 19.06.2003 |
Authors: Leitner, D., Wahl, M., Labudde, D., Diehl, A., Schmieder, P., Pires, J.R., Fossi, M., Leidert, M., Krause, G., Oschkinat, H. |
| Citation: D. Leitner; M. Wahl; D. Labudde; G. Krause; A. Diehl; P. Schmieder; J.R. Pires; M. Fossi; U. Wiedemann; M. Leidert; H. Oschkinat*; "The solution structure of an N-terminally truncated version of the yeast CDC24p PB1 domain shows a different beta-sheet topology"; FEBS Lett. 579, 3534-3538 (2005) | |
| 06 |
| 1EGX | Solution structure of the ENA-VAS-HOMOLOG-1 (EVH1) domain of human vasodilator-stimulated phosphoprotein (VASP) |
 deposited 17.02.2000 |
Authors: Ball, L., Kühne, R., Hoffmann, B., Hafner, A., Schmieder, P., Volkmer-Engert, R., Hof, M., Wahl, M., Schneider-Mergener, J., Walter, U., Oschkinat, H., Jarchau, T. |
| Citation: L.J. Ball; R. Kühne; P. Schmieder; B. Hoffmann; R. Volkmer-Engert; J. Schneider-Mergener; A. Häfner; M. Hof; M. Wahl; U. Walter; H. Oschkinat*; T. Jarchau; "Dual epitope recognition by the VASP EVH1 domain modulates polyproline ligand specificity and binding affinity"; EMBO J. 19, 4903–4914 (2000) | |
| 05 |
| 1QGP | NMR structure of the Z-alpha domain of ADAR1 |
 deposited 03.05.1999 |
Authors: Schade, M., Turner, C.J., Kühne, R., Schmieder, P., Lowenhaupt, K., Herbert, A., Rich, A., Oschkinat, H. |
| Citation: M. Schade; C.J. Turner; R. Kühne; P. Schmieder; K. Lowenhaupt, A. Herbert; A. Rich*; H. Oschkinat; "The solution structure of the Za domain of the human RNA editing enzyme ADAR1 reveals a prepositioned binding surface for Z-DNA"; Proc. Natl. Acad. Sci. USA 96, 12465-12470 (1999) | |
| 04 |
| top |
| 1SGG | The solution structure of the SAM domain from the receptor tyrosin kinase EPHB2 |
 deposited 08.01.1999 |
Authors: Smalla, M., Schmieder, P., Kelly, M., Ter Laak, A., Krause, G., Ball, L., Wahl, M., Bork, P., Oschkinat, H. |
| Citation: M. Smalla; P. Schmieder; M. Kelly; A. ter Laak; G. Krause; L. Ball; M. Wahl; P. Bork; H. Oschkinat*; "Solution structure of the receptor tyrosine kinase EphB2 SAM domain and identification of two distinct homotypic interaction sites"; Prot. Sci. 8, 1954-1961 (1999) | |
| 03 |
| 1RAX | RA-domain of RAL Guanosine-nucleotide Dissociation Stimulator |
 deposited 13.03.1999 |
Authors: Mueller, T.D., Handel, L., Schmieder, P., Oschkinat, H. |
| Citation: | |
| 02 |
| 2PDZ | Solution structure of the Syntrophin PDZ domain in complex with the peptide GVKESLV |
 deposited 10.12.1997 |
Authors: Schultz, J., Hoffmueller, U., Ashurst, J., Krause, G., Schmieder, P., Macias, M., Schneider-Mergener, J., Oschkinat, H. |
| Citation: J. Schultz; U. Hoffmüller; G. Krause; J. Ashurst; M.J. Macias; P. Schmieder; J. Schneider-Mergener; H. Oschkinat*; "Specific interactions between the syntrophin PDZ domain and voltage-gated sodium channels"; Nat. Struc. Biol. 5, 19-24 (1998) | |
| 01 |
| 6XVT | 6HQC | 5NCP | 5NBX | 5NC2 | |||||
| 5NC7 | 5NCG | 5ND0 | 5NDU | 5NEG | 5OF1 | 5OF2 | 5N91 | 5N9C | 5N9P |
| 5NCF | 5NAJ | 5NBF | 5IB1 | 5IB2 | 5IB3 | 5IB4 | 5IB5 | 5DR5 | 4MY6 |
| 6XVT | ENAH EVH1 in complex with Ac-[2-Cl-F]-PPPPTEDDL-NH2 |
| 6HQC | Structural investigation of the TasA anchoring protein TapA from Bacillus subtilis |
| 5NCP | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-12]-OH |
| 5NBX | ENAH EVH1 in complex with Ac-[2-Cl-F]-PP-[ProM-9]-OH |
| 5NC2 | ENAH EVH1 in complex with Ac-[2-Cl-F]-PPPPTEDEL-NH2 |
| 5NC7 | ENAH EVH1 in complex with Ac-WPPPPTEDEL-NH2 |
| 5NCG | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-9]-OH |
| 5NDO | ENAH EVH1 in complex with Ac-[2-Cl-F]-PP-[ProM-1]-TEDEL-NH2 |
| 5NDU | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-12]-OMe |
| 5NEG | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-13]-OEt |
| 5OF1 | The structural versatility of TasA in B. subtilis biofilm formation |
| 5OF2 | The structural versatility of TasA in B. subtilis biofilm formation |
| 5N91 | ENAH EVH1 in complex with Ac-[2-Cl-F]-PPPP-OH |
| 5N9C | ENAH EVH1 in complex with Ac-[2-Cl-F]-PP-[ProM-1]-OH |
| 5N9P | ENAH EVH1 in complex with Ac-[2-Cl-F]-PP-[ProM-1]-NH2 |
| 5NCF | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-4]-OH |
| 5NAJ | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-1]-[ProM-1]-OH |
| 5NBF | ENAH EVH1 in complex with Ac-[2-Cl-F]-[ProM-2]-[ProM-3]-OH |
| 5IB1 | Crystal structure of HLA-B*27:05 complexed with the self-peptide pVIPR measured at 295 K |
| 5IB2 | Crystal structure of HLA-B*27:05 complexed with the self-peptide pVIPR |
| 5IB3 | Crystal structure of HLA-B*27:05 complexed with the self-peptide pVIPR and Copper |
| 5IB4 | Crystal structure of HLA-B*27:05 complexed with the self-peptide pVIPR and Nickel |
| 5IB5 | Crystal structure of HLA-B*27:09 complexed with the self-peptide pVIPR and Copper |
| 5DR5 | Crystal structure of the sclerostin-neutralizing Fab AbD09097 |
| 4MY6 | EnaH-EVH1 in complex with peptidomimetic low-molecular weight inhibitor Ac-[2-Cl-F]-[ProM-2]-[ProM-1]-OH |
 |
last changes: 02.04.20, Peter Schmieder |
||